As many cheminformaticians know that (I expected…) REINVENT which is developed by AZ team is one of the useful and famous AI based compound generator in cheminformatics field. New version of REINVENT 4 is still active and recently it is version apped and added some useful code on the repositly.
The DL framework is moved from pytorch 1.x to 2.x and cool example notebooks are available.
https://github.com/MolecularAI/REINVENT4
I tried to use it today ;)
If you’ve instaled reinvent already, you should update it before run the new code. The way to update is described in README.md.
#Updating dependencies
#Update the lock files with pip-tools (please, do not edit the files #manually):
pip-compile --extra-index-url=https://download.pytorch.org/whl/cu121 --extra-index-url=https://pypi.anaconda.org/OpenEye/simple --resolver=backtracking pyproject.toml
I modified original code in my env so, I could not update code smoothly so I remove my old env and create new env again.
Then I installed some packages to run the new code.
pip install jupytext mols2grid seaborn
After the installation I tried to run the example code. Following code is same as original README.md so there are nothing to get new thing from the blog post ;P
The new version of reinvent has notebooks directory.
(reinvent4)$ cd REINVENT4/notebooks
#generate jupyter notebook from .py file with jupytext.
(reinvent4)$ jupytext --to ipynb -o Reinvent_TLRL.ipynb Reinvent_TLRL.py
After running the code above, I got ipynb file. The notebook is really cool because the code uses mol2grid and tensorboad on the notebook and provides nice view.
The code has 2 step Reinforcement Learning process.
1step: learn drug like molecules from reinvent.prior model. The drug likeness is defined with QED and alert sturcture.
2step: Target (ankyrase-2 IC50 data is used) focused feature learning with chemprop QSAR model.
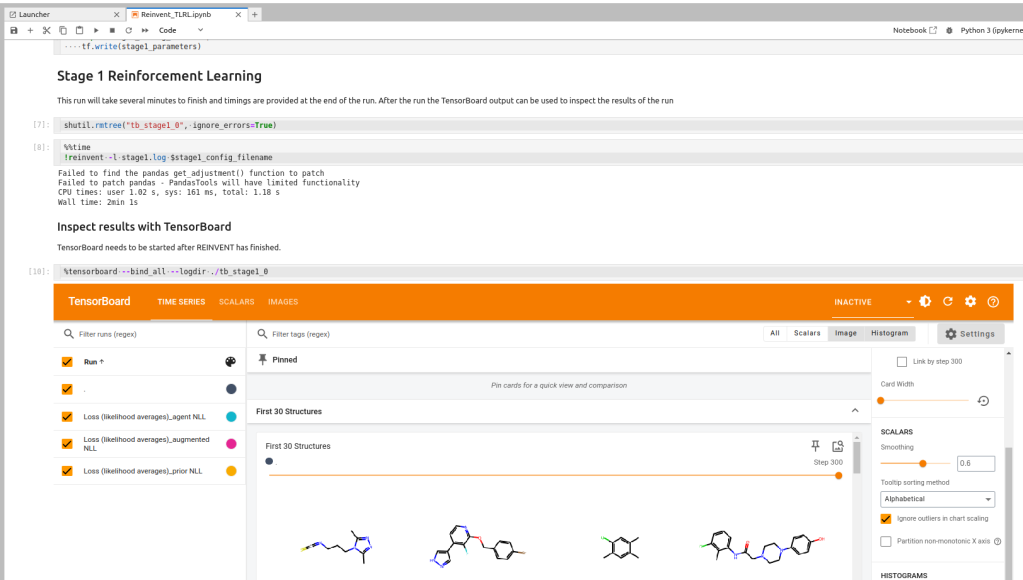
Here is the screen shot of 1st step. I didn’t know that tensorboard can render in notebook!
After the first step, transfer learning is conducted with tankylase dataset.
Also I could get cool view on the notebook. Here is a grid view of active molecules.

And learing curve from tensorboard.
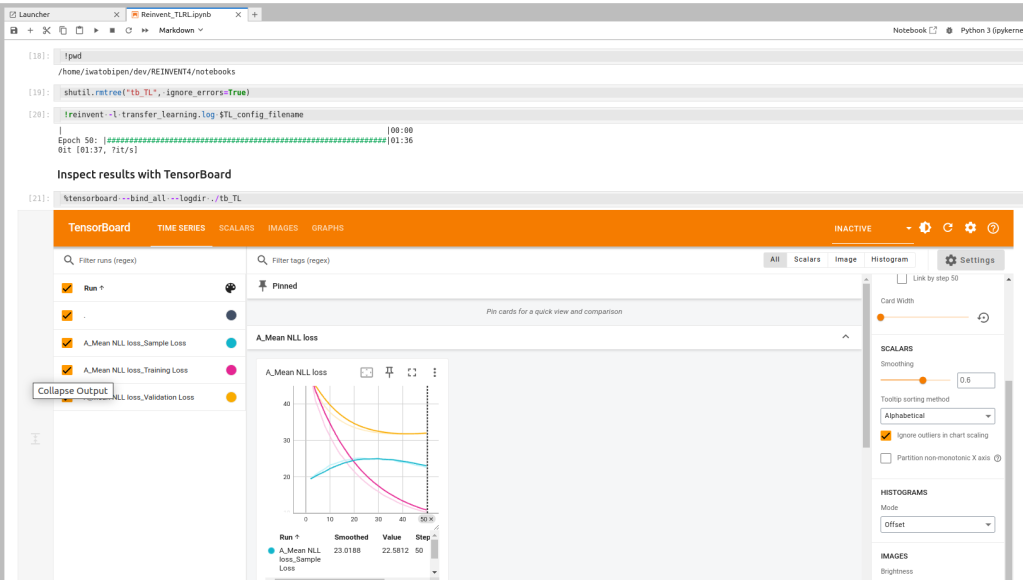
The example is useful to know that how to use REINVET4 with prior models. The example code provides not only how to run reinvent but also the discussion of eash learning step. It worth to learn for me.
I would like to say thank to the great work. And I recommend to update REINVENT in your env!
BTW, there are lots of compound generative models. And these models can generate chemically reasonable compounds. I think it’s really amazing! But still needed to deep discussion with medicinal chemists. I would like to discuss cheminformatician who uses generative models how to use them in real drug discovery projects.
Any comments, discussions are greatly appreciated.
Hi, thanks for putting this out. I just installed REINVENT4 but have not run it yet. I am also looking at SyntheMol, should be of interest for you also:
https://github.com/swansonk14/SyntheMol
Best wishes/Evert
Thanks for your information!
I’ll check it ;)
Unfortunately the REINVENT4 code does not run on my machine, and I am getting very frustrated with open-source code lately…
some contents duplicated ?
Thanks! I fixed it.