Designing linked molecule from fragments is one of the important task for drug desing such as FBDD, Scaffold hopping (e.g. replace core) and PROTAC molecule design.
As readers know there are lots of solutions to do it, for examoke BROOD is one of the famous commercial package for fragment replacement. I can’t use commercial package in my hobby so I like OSS which can apply for compound design ;)
Today I would like to share difflinker which can design linker with diffusion model. The original article is open access, I would like to share the URL.
https://www.nature.com/articles/s42256-024-00815-9
The author shares the code on github. So I tried to use it.
https://github.com/igashov/DiffLinker
The intesting point for me is that DiffLinker can link more than two fragments and can set pocket constrain. It means that we can design linker which fit given pocket constrains. It is cool isn’t it.
Okey, let’s try the code!
At first I made env for difflinker.
$ gh repo clone igashov/DiffLinker
$ cd DiffLinker
$ mamba create -c conda-forge -n difflinker rdkit
$ conda activate difflinker
$ mamba install -c conda-forge -c pytorch biopython imageio networkx pytorch pytorch-lightning scipy scikit-learn tqdm wandb
I don’t recommend making env with yml file which is included repo because the version of pytorch is little bit old so the env can’t work on recent GPUs.
After making the env, I got model parameters. The details are described on README.md
mkdir -p models
wget https://zenodo.org/record/7121300/files/geom_difflinker.ckpt?download=1 -O models/geom_difflinker.ckpt
wget https://zenodo.org/record/7121300/files/geom_size_gnn.ckpt?download=1 -O models/geom_size_gnn.ckpt
wget https://zenodo.org/records/10988017/files/pockets_difflinker_full_no_anchors_fc_pdb_excluded.ckpt?download=1 -O models/pockets_difflinker_full.ckpt
wget https://zenodo.org/records/10988017/files/pockets_difflinker_full_no_anchors_fc_pdb_excluded.ckpt?download=1 -O models/pockets_difflinker_full.ckpt
wget https://zenodo.org/record/7121300/files/pockets_difflinker_backbone.ckpt?download=1 -O models/pockets_difflinker_backbone.ckpt
wget https://zenodo.org/records/10988017/files/pockets_difflinker_full_fc_pdb_excluded.ckpt?download=1 -O models/pockets_difflinker_full_given_anchors.ckpt
wget https://zenodo.org/record/7121300/files/zinc_size_gnn.ckpt?download=1 -O models/zinc_size_gnn.ckpt
Now almost there ;) I tried by using COT kinase PDB for example.
I removed central core from the ligand to make input fragment file shown below.

Then I run the script with following commad.
$ python generate_with_protein.py --protein testdata/4y85_apo.pdb --fragments testdata/ligand_frag.sdf --output sample_size5 --linker_size 5 --model models/pockets_difflinker_full.ckpt
After running the code, I could get generated molecules with xyz format in sample_size5 directory. Then I converted the xyz file to mol format with openbabel.
$ cd sample_size5
$ $ obabel -m -ixyz *.xyz -omol
After that I could get mol files of generated molecules. Here is a result.
Gray molecule which located left bottom is original ligand and the others are generated ligans.
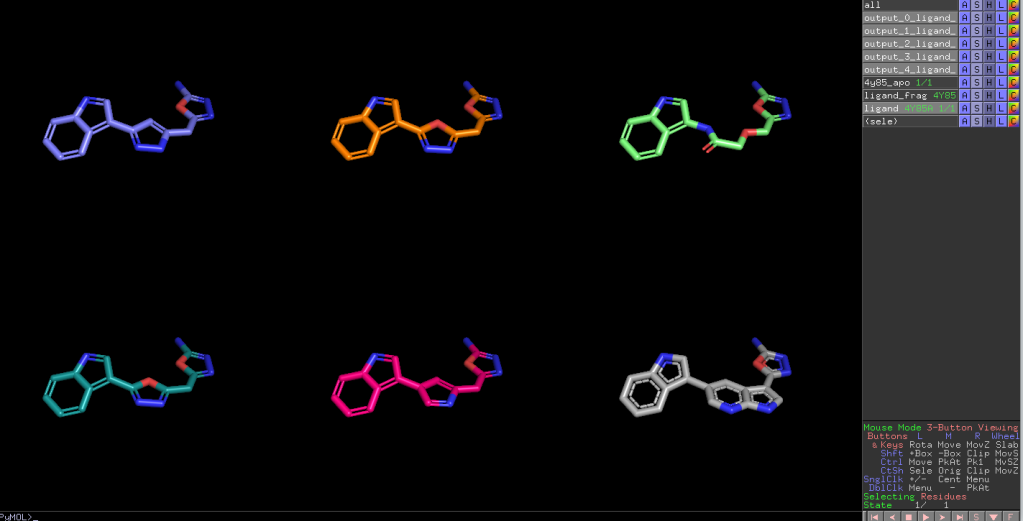
Overlayed image is shown below.

As you can see all ligands are well aligned. In summary DiffLinker is reall interesting tool for fragment linking.
I would like to generate lots of molecules with the package. Thanks for sharing such as cool code!