Atom indecies are unique number of each atom. And RDKit adds the index when mol object is generated. RDKit makes mol object from SMILES, Inchi, molblock and lots of formats. To make canonical representation of molecules, the atom indices are asigned allways same roules in automatically.
The indices are asigned regardless of scaffold. So if you have two molecules which have different substituents and same scaffold will not have same indices on the scaffold. To memorize the indices atom map numbers are used sometime but I would like to know is it possible to do same thing with atom indices, it means that how to modify the atom indices of rdkit mol object.
After googling, I found useful discussion on github.
https://github.com/rdkit/rdkit/discussions/5091
rdmolops.RenumberAtoms returns new molecules with user defined order of atom indices. Here is an example, as you can see after calling the function I could get new molecule with diffrent index order.
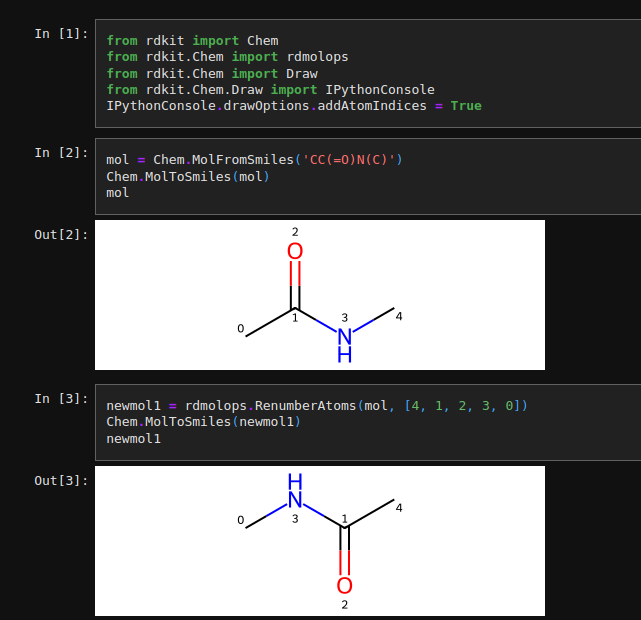
It’s worth to know that how to modify atom index on rdkit ;)
I tested the function, the code is shown on my gist.
TIPS: To get _smilesAtomOutputOrder from mol object, MolToSmiles should be called at first.
https://gist.github.com/iwatobipen/81a1c28e376fd5fc144c61c5101769b3
Thanks for reading!