Diffusion model is the one of hot area of generative model. It’s not only computer vision but also cheminformatics. Diffusion model is interesting because it generates object from some noise.
BTW, de novo compound design with target protein structure information is really attractive but difficult approach in drug design. There are some approaches to conduct it with diffusion model such as DiffSBDD. DiffSBDD generates molecule with pockect information but sometime generates strange molecules. One reason of it is DiffSBDD generates whole molecule from diffusion process without any information of scaffold. But it doesn’t represent realystic case. Because in the drug discovery project, medicinal chemists decorates molecules beased on their own scaffold. So de novo drug desing with scaffold constrain is reasonable approach.
Recently I found and read really interesting article about ‘DiffDeck’. It can get from arixv.
https://www.biorxiv.org/content/10.1101/2023.10.08.561377v1.full.pdf
Interesting points of DiffDec is listed below.
1. DiffDec can generate molecules with scaffould constrain.
2. DiffDec can generate molecule with target pocket information.
Fortunately the authors shared their code on github ‘https://github.com/biomed-AI/DiffDec‘.
I tried to use it on my PC :)
Ok, let’s dive to code! I build environment wit environment.yaml. But my pc env did not fit cuda-10.x, so I reinstalled pytorch which has cuda 11 env. But installation process is almost same as readme.md.
$ gh repo clone biomed-AI/DiffDec
$ cd DiffDec
$ mamba env create -f environment.yaml
$ conda activate DiffDec
# following package doesn't contain yaml but require.
# openbabel installation is important if the process is skipped you can't get novel molecules.
$ mamba install -c conda-forge openbabel pymol-open-source
After installation process is finished, I downloaded model parameters to run the code. The details are described in following file.
https://github.com/biomed-AI/DiffDec/blob/master/README.md
https://github.com/biomed-AI/DiffDec/blob/master/data/README.md
After putting model parametes in appropriate place, I run the code with PDB file of balicitinib which is jak2 and inhibitor complex.
DiffDec can decorate scaffold with pocket information so the molecule expect to fit target protein. To run the code, user should probide template and scaffold informatin which would like to keep in generation process. I defined scaffold of balicinib shown below.

And I prepared apo pdb and ligand pdb file from 6vn8.
Almost there, I run following command.
CUDA_VISIBLE_DEVICES=0 python sample_single_for_specific_context.py \
--scaffold_smiles_file ./data/examples/bali_scaf.smi \
--protein_file ./data/examples/jak2apo.pdb \
--scaffold_file ./data/examples/balicitinib.sdf \
--task_name exp \
--data_dir ./data/examples \
--checkpoint ./ckpt/diffdec_single.ckpt \
--samples_dir samples_jak2 \
--n_samples 50 \
--device cuda:0
After running the code, I could get new molecules with my defined scaffold.
Original structure.

Generated structure examples..


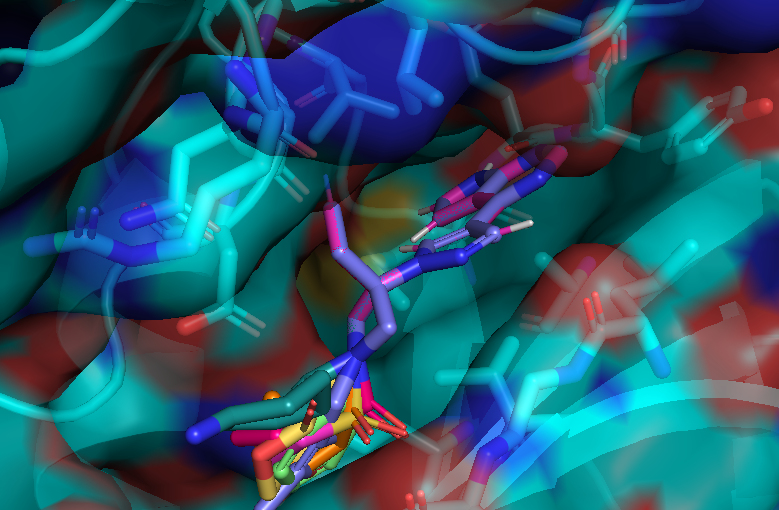
All generated molecules keep scaffold!
As arthors mentiond that this program has some limitations but it really interesting for novel compound design. I would like to use and convince other in silico approach to design new molecules.