If target structure of the drug discovery project is available, SBDD is one of the practical approach. Docking study is common method of SBDD.
There are lots of docking simulation softwares recently not only commercial packages but also open source software. AutoDock Vina is one of the famous Docking tool in the area.
To run the docking study, data preparation from receptor and ligands is required. For example in ligand preparation, several steps are requred, 3D conformation generation, predict protonation state, enumerate tautomer (if requred).
After preparation docking study is conducted with docking software. Most of the process is conducted step by step. But it will be nice if we can run the process in continuos work flow.
Fortunately I found cool package in AZ’s MoleculeAI github site which called maize.
From the original documentation,
“maize is a graph-based workflow manager for computational chemistry pipelines. It is based on the principles of flow-based programming and thus allows arbitrary graph topologies, including cycles, to be executed. Each task in the workflow (referred to as nodes) is run as a separate process and interacts with other nodes in the graph by communicating through unidirectional channels, connected to ports on each node. Every node can have an arbitrary number of input or output ports, and can read from them at any time, any number of times. This allows complex task dependencies and cycles to be modelled effectively.“
Maize can make cheminformatics pipeline very conveniently like luige. So I tried to build docking work flow with Maize today ;)
Let’s go to code!
At first, I make my conda env.
$ gh repo clone MolecularAI/maize-contrib
$ conda env create -f env-users.yml
$ conda activate maize
$ pip install --no-deps ./
Additionaly I make another env for ligand preparation. Gypsum is useful OSS package. You can read details in original article.
$ gh repo clone jcheminform/gypsum_dl
$ mamba create -n gypsum python==3.10
$ conda activate gypsum
# don't install mpi4py because it will cause error during the SBDD pipeline
$ mamba install -c conda-forge rdkit numpy scipy
All environment set up is done! Next, I made docking workflow.
The code is shown below. Most of the following code is borrowed from maize’s documentation.
First step of maize flow object is created.
import pandas as pd
from pathlib import Path
from maize.core.workflow import Workflow
from maize.steps.io import LoadData, LogResult, Return
from maize.steps.mai.docking.adv import Vina
from maize.steps.mai.molecule import Gypsum
from maize.utilities.chem import IsomerCollection
import os, sys
os.environ['XDG_CONFIG_HOME'] = '/home/iwatobipen/dev/sandbox/maizetest/'
flow = Workflow(name='dock', level='info', cleanup_temp=False)
Then load setting which tells gypsum execution info.
flow.config.update(Path("maize.toml"))
# 3D conf gen and protonate
embe = flow.add(Gypsum)
# run vina docking
dock = flow.add(Vina)
# receive docke results
retu = flow.add(Return[list[IsomerCollection]])
###maize.toml
[vina]
python = "/home/iwatobipen/miniforge3/envs/maize/bin/python"
commands.vina = "/opt/vina/bin/vina"
[gypsum]
scripts.gypsum.interpreter = "/home/iwatobipen/miniforge3/envs/gypsum/bin/python"
scripts.gypsum.location = "/home/iwatobipen/dev/gypsum_dl/run_gypsum_dl.py"
Next, read ligands as SMILES.
df = pd.read_csv('./RIPK3_chembl.csv')
print(df.shape)
smiles_lst = df['smiles'].to_list()
# used first 30 ligands
smiles_lst = [s for s in smiles_lst if type(s) == str][:30]
Then load data and configure for docking
load.data.set(smiles_lst)
embe.n_variants.set(2)
# RIP3K <https://www.rcsb.org/structure/7MON>
dock.receptor.set(Path('/home/iwatobipen/dev/sandbox/maizetest/7mon.pdbqt'))
dock.search_center.set((-21.087, -20.225, -3.082))
dock.search_range.set((20.00, 20.00,20.00))
Finally connect each module.
flow.connect(load.out, embe.inp)
flow.connect(embe.out, dock.inp)
flow.connect(dock.out,retu.inp)
flow.check()
After making flow, visualize function can make visualization of the flow ;)
All most there, let’s run the flow.
flow.execute()
2024-05-25 14:49:17,887 | INFO | dock |
___ ___ ___ ___
/\__\ /\ \ ___ /\ \ /\ \
/::| | /::\ \ /\ \ \:\ \ /::\ \
/:|:| | /:/\:\ \ \:\ \ \:\ \ /:/\:\ \
/:/|:|__|__ /::\~\:\ \ /::\__\ \:\ \ /::\~\:\ \
/:/ |::::\__\ /:/\:\ \:\__\ __/:/\/__/ _______\:\__\ /:/\:\ \:\__\
\/__/~~/:/ / \/__\:\/:/ / /\/:/ / \::::::::/__/ \:\~\:\ \/__/
/:/ / \::/ / \::/__/ \:\~~\~~ \:\ \:\__\
/:/ / /:/ / \:\__\ \:\ \ \:\ \/__/
/:/ / /:/ / \/__/ \:\__\ \:\__\
\/__/ \/__/ \/__/ \/__/
2024-05-25 14:49:17,888 | INFO | dock | Starting Maize version 0.7.0 (c) AstraZeneca 2024
2024-05-25 14:49:17,889 | INFO | dock | Running workflow 'dock' with parameters:
2024-05-25 14:49:17,890 | INFO | dock | data = ['CN1CCN(c2ccc3nc(-c4cccc(-c5cccs5)c4)c(-c4ccncc4)n3n2)CC1', 'CN1CCN(c2ccc3nc(-c4cccc(-c5ccsc5)c4)c(-c4ccncc4)n3n2)CC1', 'CN1CCN(c2ccc3nc(-c4cccc(-c5ccccc5)c4)c(-c4ccncc4)n3n2)CC1', 'CN1CCN(c2ccc3nc(-c4cccc5ccccc45)c(-c4ccncc4)n3n2)CC1', 'COc1ccc2c(-c3nc4ccc(N5CCN(C)CC5)nn4c3-c3ccncc3)cccc2c1', 'CCOc1ccc2c(-c3nc4ccc(N5CCN(C)CC5)nn4c3-c3ccncc3)cccc2c1', 'CN1CCN(c2ccc3nc(-c4cccc5c4OCO5)c(-c4ccncc4)n3n2)CC1', 'c1cc2c(c(-c3nc4cccnn4c3-c3ccncc3)c1)OCO2', 'CN1CCN(c2ccc3nc(-c4ccc5c(c4)OCO5)c(-c4ccncc4)n3n2)CC1', 'CN1CCN(c2ccc3nc(-c4ccc5ncsc5c4)c(-c4ccncc4)n3n2)CC1', 'CN1CCN(c2ccc3nc(-c4cccc5c4OCO5)c(-c4ccncc4)n3c2)CC1', 'CN1CCN(c2ccc3nc(-c4cccc5c4OCO5)c(-c4ccccc4)n3n2)CC1', 'CN1CCN(c2ccc3nc(-c4ccc5c(c4)OCCO5)c(-c4ccncc4)n3n2)CC1', 'CN1CCN(c2ccc3nc(-c4cccc5c4OCCO5)c(-c4ccncc4)n3n2)CC1', 'CN1CCN(c2ccc3nc(-c4cccc(C(=O)N5CCOCC5)c4)c(-c4ccncc4)n3n2)CC1', 'CN1CCN(c2ccc3nc(-c4ccc(-c5ccccc5)cc4)c(-c4ccncc4)n3n2)CC1', 'COc1cccc(-c2nc3ccc(N4CCN(C)CC4)nn3c2-c2ccncc2)c1', 'CN1CCN(c2ccc3nc(-c4ccc(C(=O)N5CCOCC5)cc4)c(-c4ccncc4)n3n2)CC1', 'CN1CCN(c2ccc3nc(-c4ccc(-c5ccccc5)s4)c(-c4ccncc4)n3n2)CC1', 'CN1CCN(c2ccc3nc(-c4cccc5c4OCO5)c(-c4cccnc4)n3n2)CC1', 'Cn1c(=O)c(S(=O)(=O)c2ccc(F)cc2F)cc2cnc(Nc3ccc4[nH]ccc4c3)nc21', 'CC(C)(C)S(=O)(=O)c1ccc2nccc(Nc3n[nH]c4ccc(F)cc34)c2c1', 'Cc1cc(-c2cc(O)ccc2Cl)cc2nnc(Nc3ccc(OCCN4CCCC4)cc3)nc12', 'Cc1cccc(-c2nn3c(c2-c2ccnc4ccc(C(N)=O)cc24)CCC3)n1', 'Cc1ccc(C(=O)Nc2ccc(CN3CCN(C)CC3)c(C(F)(F)F)c2)cc1C#Cc1cnc2cccnn12', 'C=CC(=O)N1CCC[C@@H](n2nc(-c3ccc(Oc4ccccc4)cc3)c3c(N)ncnc32)C1', 'C[C@@H]1COC(Nc2ccc3ncnc(Nc4ccc(OCc5nccs5)c(Cl)c4)c3c2)=N1', 'COc1cc2c(Oc3ccc(NC(=O)C4(C(=O)Nc5ccc(F)cc5)CC4)cc3F)ccnc2cc1OCCCN1CCOCC1', 'CN1CCN(C2CCN(C(=O)Nc3cc(Oc4ccc(NC(=O)C5(C(=O)Nc6ccc(F)cc6)CC5)c(F)c4)ccn3)CC2)CC1', 'COc1cc2ncnc(Oc3cccc(NC(=O)Nc4cc(C(C)(C)C(F)(F)F)on4)c3)c2cc1OC'] (from 'loaddata')
2024-05-25 14:49:17,890 | INFO | dock | n_variants = 2 (from 'gypsum')
2024-05-25 14:49:17,891 | INFO | dock | receptor = /home/iwatobipen/dev/sandbox/maizetest/7mon.pdbqt (from 'vina')
2024-05-25 14:49:17,891 | INFO | dock | search_center = (-21.087, -20.225, -3.082) (from 'vina')
2024-05-25 14:49:17,892 | INFO | dock | search_range = (20.0, 20.0, 20.0) (from 'vina')
2024-05-25 14:49:18,606 | INFO | dock | Node 'loaddata' finished (1/4)
\
2024-05-25 14:49:32,804 | INFO | dock | Workflow status
| loaddata | COMPLETED
| vina | WAITING_FOR_INPUT
| return | WAITING_FOR_INPUT
| gypsum | RUNNING
2024-05-25 14:49:32,804 | INFO | gypsum | Found failed SMILES file
2024-05-25 14:49:32,804 | INFO | gypsum | Failed SMILES:
'CN1CCN(C2CCN(C(=O)Nc3cc(Oc4ccc(NC(=O)C5(C(=O)Nc6ccc(F)cc6)CC5)c(F)c4)ccn3)CC2)CC1
C=CC(=O)N1CCC[C@@H](n2nc(-c3ccc(Oc4ccccc4)cc3)c3c(N)ncnc32)C1'
2024-05-25 14:49:32,854 | WARNING | gypsum | Skipping failed embedding for SMILES 'C=CC(=O)N1CCC[C@@H](n2nc(-c3ccc(Oc4ccccc4)cc3)c3c(N)ncnc32)C1', falling back to RDKit
2024-05-25 14:49:32,934 | WARNING | gypsum | Skipping failed embedding for SMILES 'CN1CCN(C2CCN(C(=O)Nc3cc(Oc4ccc(NC(=O)C5(C(=O)Nc6ccc(F)cc6)CC5)c(F)c4)ccn3)CC2)CC1', falling back to RDKit
2024-05-25 14:49:33,067 | INFO | vina | Docking molecule 0: 'IsomerCollection('CN1CCN(c2ccc3nc(-c4cccc(-c5cccs5)c4)c(-c4ccncc4)n3n2)CC1', n_isomers=2)'
2024-05-25 14:49:33,078 | INFO | vina | Docking molecule 1: 'IsomerCollection('CN1CCN(c2ccc3nc(-c4cccc(-c5ccsc5)c4)c(-c4ccncc4)n3n2)CC1', n_isomers=2)'
2024-05-25 14:49:33,093 | INFO | vina | Docking molecule 2: 'IsomerCollection('CN1CCN(c2ccc3nc(-c4cccc(-c5ccccc5)c4)c(-c4ccncc4)n3n2)CC1', n_isomers=2)'
2024-05-25 14:49:33,158 | INFO | vina | Docking molecule 3: 'IsomerCollection('CN1CCN(c2ccc3nc(-c4cccc5ccccc45)c(-c4ccncc4)n3n2)CC1', n_isomers=2)'
2024-05-25 14:49:33,169 | INFO | vina | Docking molecule 4: 'IsomerCollection('COc1ccc2c(-c3nc4ccc(N5CCN(C)CC5)nn4c3-c3ccncc3)cccc2c1', n_isomers=2)'
2024-05-25 14:49:33,180 | INFO | vina | Docking molecule 5: 'IsomerCollection('CCOc1ccc2c(-c3nc4ccc(N5CCN(C)CC5)nn4c3-c3ccncc3)cccc2c1', n_isomers=2)'
2024-05-25 14:49:33,190 | INFO | vina | Docking molecule 6: 'IsomerCollection('CN1CCN(c2ccc3nc(-c4cccc5c4OCO5)c(-c4ccncc4)n3n2)CC1', n_isomers=2)'
2024-05-25 14:49:33,200 | INFO | vina | Docking molecule 7: 'IsomerCollection('c1cc2c(c(-c3nc4cccnn4c3-c3ccncc3)c1)OCO2', n_isomers=2)'
|
2024-05-25 14:49:33,207 | INFO | vina | Docking molecule 8: 'IsomerCollection('CN1CCN(c2ccc3nc(-c4ccc5c(c4)OCO5)c(-c4ccncc4)n3n2)CC1', n_isomers=2)'
2024-05-25 14:49:33,217 | INFO | vina | Docking molecule 9: 'IsomerCollection('CN1CCN(c2ccc3nc(-c4ccc5ncsc5c4)c(-c4ccncc4)n3n2)CC1', n_isomers=2)'
2024-05-25 14:49:33,226 | INFO | vina | Docking molecule 10: 'IsomerCollection('CN1CCN(c2ccc3nc(-c4cccc5c4OCO5)c(-c4ccncc4)n3c2)CC1', n_isomers=2)'
2024-05-25 14:49:33,235 | INFO | vina | Docking molecule 11: 'IsomerCollection('CN1CCN(c2ccc3nc(-c4cccc5c4OCO5)c(-c4ccccc4)n3n2)CC1', n_isomers=2)'
2024-05-25 14:49:33,244 | INFO | vina | Docking molecule 12: 'IsomerCollection('CN1CCN(c2ccc3nc(-c4ccc5c(c4)OCCO5)c(-c4ccncc4)n3n2)CC1', n_isomers=2)'
2024-05-25 14:49:33,254 | INFO | vina | Docking molecule 13: 'IsomerCollection('CN1CCN(c2ccc3nc(-c4cccc5c4OCCO5)c(-c4ccncc4)n3n2)CC1', n_isomers=2)'
2024-05-25 14:49:33,264 | INFO | vina | Docking molecule 14: 'IsomerCollection('CN1CCN(c2ccc3nc(-c4cccc(C(=O)N5CCOCC5)c4)c(-c4ccncc4)n3n2)CC1', n_isomers=2)'
2024-05-25 14:49:33,274 | INFO | vina | Docking molecule 15: 'IsomerCollection('CN1CCN(c2ccc3nc(-c4ccc(-c5ccccc5)cc4)c(-c4ccncc4)n3n2)CC1', n_isomers=2)'
2024-05-25 14:49:33,284 | INFO | vina | Docking molecule 16: 'IsomerCollection('COc1cccc(-c2nc3ccc(N4CCN(C)CC4)nn3c2-c2ccncc2)c1', n_isomers=2)'
2024-05-25 14:49:33,293 | INFO | vina | Docking molecule 17: 'IsomerCollection('CN1CCN(c2ccc3nc(-c4ccc(C(=O)N5CCOCC5)cc4)c(-c4ccncc4)n3n2)CC1', n_isomers=2)'
2024-05-25 14:49:33,304 | INFO | vina | Docking molecule 18: 'IsomerCollection('CN1CCN(c2ccc3nc(-c4ccc(-c5ccccc5)s4)c(-c4ccncc4)n3n2)CC1', n_isomers=2)'
2024-05-25 14:49:33,314 | INFO | vina | Docking molecule 19: 'IsomerCollection('CN1CCN(c2ccc3nc(-c4cccc5c4OCO5)c(-c4cccnc4)n3n2)CC1', n_isomers=2)'
2024-05-25 14:49:33,323 | INFO | vina | Docking molecule 20: 'IsomerCollection('Cn1c(=O)c(S(=O)(=O)c2ccc(F)cc2F)cc2cnc(Nc3ccc4[nH]ccc4c3)nc21', n_isomers=2)'
2024-05-25 14:49:33,332 | INFO | vina | Docking molecule 21: 'IsomerCollection('CC(C)(C)S(=O)(=O)c1ccc2nccc(Nc3n[nH]c4ccc(F)cc34)c2c1', n_isomers=1)'
2024-05-25 14:49:33,336 | INFO | vina | Docking molecule 22: 'IsomerCollection('Cc1cc(-c2cc(O)ccc2Cl)cc2nnc(Nc3ccc(OCCN4CCCC4)cc3)nc12', n_isomers=2)'
2024-05-25 14:49:33,346 | INFO | vina | Docking molecule 23: 'IsomerCollection('Cc1cccc(-c2nn3c(c2-c2ccnc4ccc(C(N)=O)cc24)CCC3)n1', n_isomers=2)'
2024-05-25 14:49:33,354 | INFO | vina | Docking molecule 24: 'IsomerCollection('Cc1ccc(C(=O)Nc2ccc(CN3CCN(C)CC3)c(C(F)(F)F)c2)cc1C#Cc1cnc2cccnn12', n_isomers=2)'
2024-05-25 14:49:33,365 | INFO | vina | Docking molecule 25: 'IsomerCollection('C=CC(=O)N1CCC[C@@H](n2nc(-c3ccc(Oc4ccccc4)cc3)c3c(N)ncnc32)C1', n_isomers=1)'
2024-05-25 14:49:33,370 | INFO | vina | Docking molecule 26: 'IsomerCollection('C[C@@H]1COC(Nc2ccc3ncnc(Nc4ccc(OCc5nccs5)c(Cl)c4)c3c2)=N1', n_isomers=2)'
2024-05-25 14:49:33,379 | INFO | vina | Docking molecule 27: 'IsomerCollection('COc1cc2c(Oc3ccc(NC(=O)C4(C(=O)Nc5ccc(F)cc5)CC4)cc3F)ccnc2cc1OCCCN1CCOCC1', n_isomers=2)'
2024-05-25 14:49:33,392 | INFO | vina | Docking molecule 28: 'IsomerCollection('CN1CCN(C2CCN(C(=O)Nc3cc(Oc4ccc(NC(=O)C5(C(=O)Nc6ccc(F)cc6)CC5)c(F)c4)ccn3)CC2)CC1', n_isomers=1)'
2024-05-25 14:49:33,399 | INFO | vina | Docking molecule 29: 'IsomerCollection('COc1cc2ncnc(Oc3cccc(NC(=O)Nc4cc(C(C)(C)C(F)(F)F)on4)c3)c2cc1OC', n_isomers=2)'
2024-05-25 14:49:33,555 | INFO | dock | Node 'gypsum' finished (2/4)
\
2024-05-25 14:49:54,600 | INFO | vina | Job completed (0/57)
|
2024-05-25 14:49:58,247 | INFO | vina | Job completed (1/57)
\
2024-05-25 14:50:18,508 | INFO | vina | Job completed (2/57)
2024-05-25 14:50:20,215 | INFO | vina | Job completed (3/57)
|
2024-05-25 14:50:21,526 | INFO | vina | Job completed (4/57)
-
2024-05-25 14:50:27,853 | INFO | vina | Job completed (5/57)
2024-05-25 14:50:28,143 | INFO | vina | Job completed (6/57)
2024-05-25 14:50:28,585 | INFO | vina | Job completed (7/57)
\
2024-05-25 14:50:30,746 | INFO | vina | Job completed (8/57)
/
2024-05-25 14:50:38,254 | INFO | vina | Job completed (9/57)
2024-05-25 14:50:38,996 | INFO | vina | Job completed (10/57)
\
2024-05-25 14:50:44,581 | INFO | vina | Job completed (11/57)
|
2024-05-25 14:50:47,193 | INFO | vina | Job completed (12/57)
/
2024-05-25 14:50:51,085 | INFO | vina | Job completed (13/57)
2024-05-25 14:50:51,096 | INFO | vina | Job completed (14/57)
-
2024-05-25 14:50:51,552 | INFO | vina | Job completed (15/57)
2024-05-25 14:50:52,134 | INFO | vina | Job completed (16/57)
\
2024-05-25 14:50:56,266 | INFO | vina | Job completed (17/57)
\
2024-05-25 14:51:06,757 | INFO | vina | Job completed (18/57)
2024-05-25 14:51:07,283 | INFO | vina | Job completed (19/57)
|
2024-05-25 14:51:09,968 | INFO | vina | Job completed (20/57)
2024-05-25 14:51:10,063 | INFO | vina | Job completed (21/57)
/
2024-05-25 14:51:15,306 | INFO | vina | Job completed (22/57)
-
2024-05-25 14:51:16,527 | INFO | vina | Job completed (23/57)
\
2024-05-25 14:51:21,211 | INFO | vina | Job completed (24/57)
\
2024-05-25 14:51:33,463 | INFO | vina | Job completed (25/57)
|
2024-05-25 14:51:34,720 | INFO | vina | Job completed (26/57)
2024-05-25 14:51:35,520 | INFO | vina | Job completed (27/57)
/
2024-05-25 14:51:38,892 | INFO | vina | Job completed (28/57)
-
2024-05-25 14:51:42,529 | INFO | vina | Job completed (29/57)
-
2024-05-25 14:51:51,864 | INFO | vina | Job completed (30/57)
/
2024-05-25 14:52:00,942 | INFO | vina | Job completed (31/57)
2024-05-25 14:52:01,757 | INFO | vina | Job completed (32/57)
2024-05-25 14:52:03,218 | INFO | vina | Job completed (33/57)
-
2024-05-25 14:52:05,319 | INFO | vina | Job completed (34/57)
\
2024-05-25 14:52:08,447 | INFO | vina | Job completed (35/57)
|
2024-05-25 14:52:10,878 | INFO | vina | Job completed (36/57)
\
2024-05-25 14:52:19,815 | INFO | vina | Job completed (37/57)
2024-05-25 14:52:20,742 | INFO | vina | Job completed (38/57)
|
2024-05-25 14:52:23,336 | INFO | vina | Job completed (39/57)
2024-05-25 14:52:24,244 | INFO | vina | Job completed (40/57)
2024-05-25 14:52:24,288 | INFO | vina | Job completed (41/57)
/
2024-05-25 14:52:26,709 | INFO | vina | Job completed (42/57)
2024-05-25 14:52:26,969 | INFO | vina | Job completed (43/57)
|
2024-05-25 14:52:34,102 | INFO | vina | Job completed (44/57)
/
2024-05-25 14:52:51,533 | INFO | vina | Job completed (45/57)
/
2024-05-25 14:53:01,733 | INFO | vina | Job completed (46/57)
2024-05-25 14:53:02,079 | INFO | vina | Job completed (47/57)
\
2024-05-25 14:53:09,269 | INFO | vina | Job completed (48/57)
|
2024-05-25 14:53:11,683 | INFO | vina | Job completed (49/57)
/
2024-05-25 14:53:15,186 | INFO | vina | Job completed (50/57)
|
2024-05-25 14:53:35,040 | INFO | vina | Job completed (51/57)
-
2024-05-25 14:53:52,067 | INFO | vina | Job completed (52/57)
/
2024-05-25 14:54:01,673 | INFO | vina | Job completed (53/57)
\
2024-05-25 14:54:20,071 | INFO | vina | Job completed (54/57)
|
2024-05-25 14:55:22,603 | INFO | vina | Job completed (55/57)
2024-05-25 14:55:23,648 | INFO | vina | Job completed (56/57)
2024-05-25 14:55:23,693 | INFO | dock | Workflow status
| loaddata | COMPLETED
| vina | RUNNING
| return | WAITING_FOR_INPUT
| gypsum | COMPLETED
2024-05-25 14:55:23,727 | INFO | vina | Parsing isomer 0: 'Isomer(n_atoms=58, n_conformers=1, charge=1)'
2024-05-25 14:55:23,739 | INFO | vina | Parsing isomer 1: 'Isomer(n_atoms=58, n_conformers=1, charge=1)'
2024-05-25 14:55:23,742 | INFO | vina | Parsing isomer 2: 'Isomer(n_atoms=61, n_conformers=1, charge=1)'
2024-05-25 14:55:23,745 | INFO | vina | Parsing isomer 3: 'Isomer(n_atoms=57, n_conformers=1, charge=1)'
2024-05-25 14:55:23,747 | INFO | vina | Parsing isomer 4: 'Isomer(n_atoms=58, n_conformers=1, charge=2)'
2024-05-25 14:55:23,750 | INFO | vina | Parsing isomer 5: 'Isomer(n_atoms=61, n_conformers=1, charge=1)'
2024-05-25 14:55:23,753 | INFO | vina | Parsing isomer 6: 'Isomer(n_atoms=61, n_conformers=1, charge=1)'
2024-05-25 14:55:23,755 | INFO | vina | Parsing isomer 7: 'Isomer(n_atoms=65, n_conformers=1, charge=2)'
2024-05-25 14:55:23,758 | INFO | vina | Parsing isomer 8: 'Isomer(n_atoms=53, n_conformers=1, charge=0)'
2024-05-25 14:55:23,760 | INFO | vina | Parsing isomer 9: 'Isomer(n_atoms=37, n_conformers=1, charge=1)'
2024-05-25 14:55:23,763 | INFO | vina | Parsing isomer 10: 'Isomer(n_atoms=54, n_conformers=1, charge=1)'
2024-05-25 14:55:23,765 | INFO | vina | Parsing isomer 11: 'Isomer(n_atoms=52, n_conformers=1, charge=0)'
2024-05-25 14:55:23,767 | INFO | vina | Parsing isomer 12: 'Isomer(n_atoms=55, n_conformers=1, charge=1)'
2024-05-25 14:55:23,770 | INFO | vina | Parsing isomer 13: 'Isomer(n_atoms=54, n_conformers=1, charge=0)'
2024-05-25 14:55:23,772 | INFO | vina | Parsing isomer 14: 'Isomer(n_atoms=56, n_conformers=1, charge=0)'
2024-05-25 14:55:23,775 | INFO | vina | Parsing isomer 15: 'Isomer(n_atoms=57, n_conformers=1, charge=1)'
2024-05-25 14:55:23,777 | INFO | vina | Parsing isomer 16: 'Isomer(n_atoms=66, n_conformers=1, charge=1)'
2024-05-25 14:55:23,780 | INFO | vina | Parsing isomer 17: 'Isomer(n_atoms=61, n_conformers=1, charge=1)'
2024-05-25 14:55:23,783 | INFO | vina | Parsing isomer 18: 'Isomer(n_atoms=54, n_conformers=1, charge=0)'
2024-05-25 14:55:23,785 | INFO | vina | Parsing isomer 19: 'Isomer(n_atoms=66, n_conformers=1, charge=1)'
2024-05-25 14:55:23,788 | INFO | vina | Parsing isomer 20: 'Isomer(n_atoms=59, n_conformers=1, charge=2)'
2024-05-25 14:55:23,790 | INFO | vina | Parsing isomer 21: 'Isomer(n_atoms=58, n_conformers=1, charge=1)'
2024-05-25 14:55:23,793 | INFO | vina | Parsing isomer 22: 'Isomer(n_atoms=54, n_conformers=1, charge=1)'
2024-05-25 14:55:23,795 | INFO | vina | Parsing isomer 23: 'Isomer(n_atoms=49, n_conformers=1, charge=1)'
2024-05-25 14:55:23,797 | INFO | vina | Parsing isomer 24: 'Isomer(n_atoms=48, n_conformers=1, charge=0)'
2024-05-25 14:55:23,800 | INFO | vina | Parsing isomer 25: 'Isomer(n_atoms=47, n_conformers=1, charge=0)'
2024-05-25 14:55:23,802 | INFO | vina | Parsing isomer 26: 'Isomer(n_atoms=60, n_conformers=1, charge=0)'
2024-05-25 14:55:23,805 | INFO | vina | Parsing isomer 27: 'Isomer(n_atoms=47, n_conformers=1, charge=0)'
2024-05-25 14:55:23,807 | INFO | vina | Parsing isomer 28: 'Isomer(n_atoms=46, n_conformers=1, charge=-1)'
2024-05-25 14:55:23,809 | INFO | vina | Parsing isomer 29: 'Isomer(n_atoms=67, n_conformers=1, charge=1)'
2024-05-25 14:55:23,812 | INFO | vina | Parsing isomer 30: 'Isomer(n_atoms=57, n_conformers=1, charge=0)'
2024-05-25 14:55:23,814 | INFO | vina | Parsing isomer 31: 'Isomer(n_atoms=52, n_conformers=1, charge=1)'
2024-05-25 14:55:23,816 | INFO | vina | Parsing isomer 32: 'Isomer(n_atoms=52, n_conformers=1, charge=1)'
2024-05-25 14:55:23,819 | INFO | vina | Parsing isomer 33: 'Isomer(n_atoms=80, n_conformers=1, charge=0)'
2024-05-25 14:55:23,822 | INFO | vina | Parsing isomer 34: 'Isomer(n_atoms=83, n_conformers=1, charge=0)'
2024-05-25 14:55:23,824 | INFO | vina | Parsing isomer 35: 'Isomer(n_atoms=58, n_conformers=1, charge=-1)'
2024-05-25 14:55:23,827 | INFO | vina | Parsing isomer 36: 'Isomer(n_atoms=58, n_conformers=1, charge=-1)'
2024-05-25 14:55:23,868 | INFO | dock | Node 'return' finished (3/4)
2024-05-25 14:55:24,343 | INFO | dock | Node 'vina' finished (4/4)
2024-05-25 14:55:24,846 | INFO | dock | Execution completed :) total runtime: 0:06:06.423945
4 nodes completed successfully
0 nodes stopped due to closing ports
0 nodes failed
0:12:26.539768 total walltime
0:06:21.405047 spent waiting for resources or other nodes
After execution, I could get docking results shown below.
res=retu.get()
import numpy as np
from pathlib import Path
for idx, m in enumerate(res):
if type(m.best_score)==np.float64:
m.to_sdf(Path(f'ok_{idx}.sdf'))
else:
# docking failed compounds don't have score.
m.to_sdf(Path(f'ng_{idx}.sdf'))
After running the final code, I could get sdf of docked compounds.
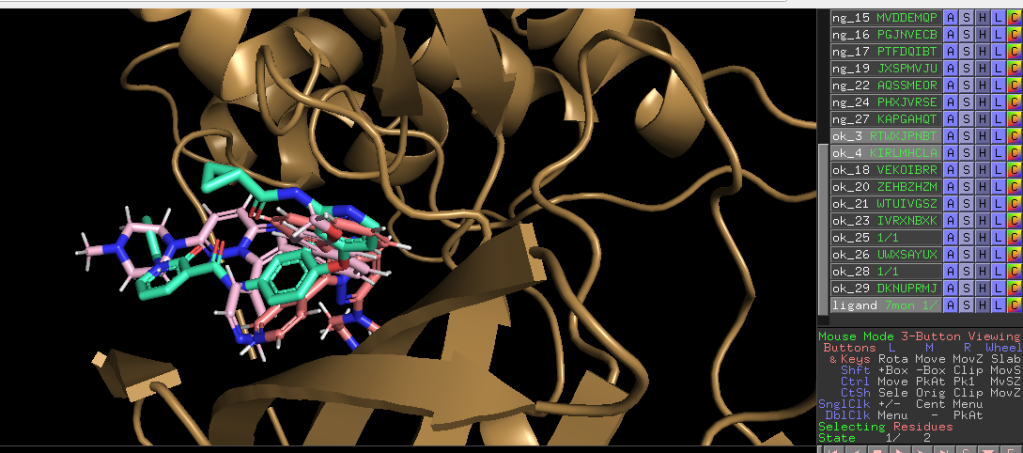
Sky bule is original ligand and the others are ligands from SMILES. Successed compound located same site as original ligand.
Maize supports several docking software vina, vina-gpu, glide etc. And also it supports not only docking but also LBDD tools such as ROCS, DL tool reinvent etc.
And also you can make your own components. It sounds nice!
In summary maize is useful package for drug discovery I would like to make my oun module near the feature.